Live Chat Log for Biology
Good morning
good morning
Good morning
I've posted the discussion question for today in the Moodle under the free exercise subject, or you can look it up at the AP Biology Website. Click on the
2007 Free-Response Questions to download the PDF with all the Free-Response questions for the 2007 exam, then look at #4.
Have you had a chance to look at the question?
yes
yes
yes
Let's see if we can make sense of the question first. We are cutting a bacterial plasmid with two different restriction enzymes in order to analyze its structure. What is a bacterial plasmid?
it is a gene dictating a certain trait
Not really. It could involve more than one gene.
its a prtion of DNA in the bacteria
A ring of DNA replicating separately from the main chromosome in the bacteria
it could even come from outside the bacteria, through transformation.
But it is a separate segment of DNA in a ring shape.
( Look back on page 205, where the concept is first introduced).
So we start with a string of DNA of hundred kb -- does anyone know what the kb stands for here?
I actually don't
Kilobase?
Good for Sarah! What's a kilobase?
a 1000 bases
Right. So we have a strand with 100 times 1000 base pairs.
We make lots of copies of it, and then we cut it with two different restriction enzymes.
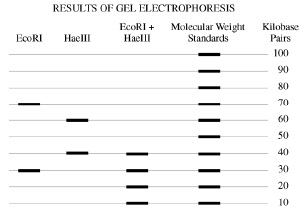
We cut it with the EcoRI restriction enzyme, we get two segments, as shown in the leftmost column of the gel electrophoresis diagram. How are the segments?
One has 70 kilobase pairs, and the other has 30
yep^
Good: here's the data -
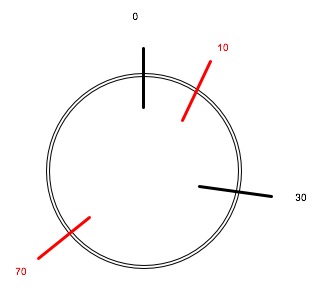
So, envision a circle divided into 100 parts, each one KB long -- that is, each contains 1000 neucleotide base pairs (A-T or C-G).
It when we use this restriction enzyme, it cuts the circle into two pieces, one 70kb long and the other 30kb long.
We want to represent this on the circle.
We arbitrarily choose a starting point at the top (this is kind of the convention) was a restriction enzyme cuts the plasmid the first time.
Then we work our way 30% (close to 33% or 1/3) of the way around the 100% of the circle in a clockwise direction. That puts us at about "four o'clock" for the second cut, since if we think of the circle is a clock face with 12 hours, one third of the distance around the clock face will be four hours.
Our circle now looks like this:
Does this make sense?
yes
On an exam, you can draw the segment break up anyway you want as long as the marks divide the circle into two 70%: 30% fragments.
Starting at the top and moving in a clockwise direction are conventions, not requirements, but it's useful to follow the conventions, since you then don't have to make a decision about which way to draw it.
do the 30% and 70% marks have to be accurate? or can they just be approximations?
In an exam situation, you won't have a way to measure around the circle, so your drawing will probably be approximate. However, you'll notice that I labeled the segments, so that for an examiner there should be no ambiguity about the lengths I meant to indicate.
Any questions on what we just did, or why?
Susan, do you have a question?
no sorry
No ma'am, I think that part is pretty straightforward
AHA..right...no, the OTHER right....
Now that you know what we're trying to do, what happens if we take a brand-new plasmid and use the HaeIII restriction enzyme to cut it?
it cuts at 40 and 60
We cut it into two parts, one 60% and the other 40%
we cut it into two parts, one containing 60% of the ring, and the other 40% of the ring
The problem is, we don't know where these are relative to the first cuts from the EcoRI that we've already indicated.
So we have to throw some new copies of the plasmids in with both restriction enzymes. What happens then?
it cuts at 10 20 30 and 40
Right. We at four separate segments: note that 10+20 +30 + 40 = 100
So the cuts are in four different places.
We need to arrange the HaeIII enzyme cuts which taken by themselves break the circle into two segments of 40kb and 60kb, and map them over the existing EcoRI cuts, which break the circle into two segments of 30kb and 70kb, and arrange them so that we wind up with four segments: 10kb, 20kb, 30kb, and 40kb.
We're going to have to use some trial and error to see what happens.
We know that one of the segments is only 10kb long.
So the cut that the HaeIII restriction enzyme makes his 10kb to either side of one of the existing EcoRI cuts.
Let's start by seeing whether we can get this to work if we put the first HaeIII cut 10 KB around the circle from the top where the first EcoRi cut is (at about "one o'clock").
This cuts the existing 30kb into two segments: one 10kb and one 20kb long.
So far, that's promising; we know that we get segments of those two lengths when we use both restriction enzymes.
OK so far?
The second cut has to go either 40 KB around to the right or 60 KB around to the right, since we got strands of 40 and 60 using just the HaeIII enzyme.
Kf we put at 40 kb from the 10 kb position, the cut will be at 50kb. We'll have cuts at 0, 10, 30, and 50, and we'll get segments 10, 20, 20, and 50 long.
This is not all we observed from the gel electrophoresis data, so a 40kb segment counting from the 10kb position won't work.
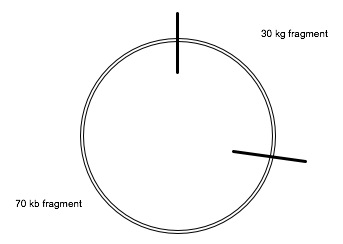
Let's try 60kb segment. That puts the second HaeIII cut at 70kb around the circle from the first EcoRI cut:
Now we have cuts at 0, 10, 30, and 70. We have segments 0-10 (10kb), 10-30 (20kb), 30-70 (40kb), and 70-100 (30kb).
That division gives us the observed DNA strand lengths, so that's how the two restriction enzymes working together cut the plasmid.
Questions?
When you do a diagram like this, be sure that you label everything.
ok
I should have labeled the two black cuts "E" or "EcoRI" and the two red cuts "H" or "HaeIII", to show that I know which restriction enzyme is making those cuts.
By the way, you can get this to work if you put the first H cut 10kb to the left (counterclockwise) from E_0, and the second H cut 40kb counterclockwise from that point.
You would then have cuts at 0(E), 30(E), 50(H), and 90(H), givine you strands of 30, 20, 40, and 10.
Since there are two legitimate answers, you would get full credit for either one.
Now let's look at rest of the question! We need to describe how the common in DNA technology could be used to insert a gene of interest into a bacterium, recombinant bacteria can be identified, an expression of the gene of interest can be ensured.
How do we go about inserting a gene or DNA sequence we are interested in into a bacterium?
Everybody stumped?
You combine the gene of interest and the plasmid, and then use DNA ligase to paste it, and then put the plasmid into the bacterium by transformation
^that sounds right to me
Good. That's the right method.
Bacteria sort of scoop up any DNA lying around, so making a soup of plasmids and tossing some bacteria in will get the new plasmid into the bacteria.
How would you know that the bacteria had picked up the plasmid?
if it starts doing something the new DNA would do, like make a certain enzyme
You could place it in an environment and compare its reactions to those of unaffected bacteria
You could clone the bacteria and then use nucleic acid probes
Either of the two methods will work: you can either look for a characteristic that could only come from the new genes, or you could use a radioactive marker (usually mRNA).
Finally, how can we be sure that the new gene will be expressed?
[ Think about what causes gene expression!]
You might want to review page 211.
put it in an environment where it would be neccissary
Okay we wanted an environment where something will trigger the expression of the gene.
In the expression of the lac operon, the existence of lactose is what turned the gene on by binding the repressor molecule so that it couldn't sit on the operator and prevent the RNA polymerase from binding to the promoter.
If we wanted genes on our designer plasmids expressed by the bacteria into which we checked them, we need to be sure to include the promoter sequence, so that the bacterium's own RNA polymerase has a binding site to start transcription of the DNA.
Makes sense?
The final part of this question asks how a genetically modified organism might be beneficial for humans and at the same time pose a threat to a population or ecosystem.
Propose a genetic modification (GM) that would both benefit humans and be a danger to non-humans or the overall ecosystem.
This is where you could come up with something either known or speculative.
Ideas?
A bacteria modified to break down oil is helpful to humans as it cleans up oil spills. This same bacteria is also poisonous, and can kill fish and other life in the area.
Good example!
they can help by makign medicines, but others might be made to decompose something and decompose to much, eatting crops and food
Just about any bacteria that produces something useful could be dangerous in the wild. If bacteria used to produce human insulin for diabetics got into the general environment, and were ingested by other animals, they would create elevated insulin levels and those animals, which would pose a serious threat to the individual animal's health.
So laboratory controls need to be very strict to prevent genetically modified bacteria from "escaping", like the rats of NIMH.
Are there any questions on any part of this free exercise?
no ma'am
I'm trying to pick examples that not only force you to practice study techniques and exam taking techniques and logical analysis, but also force you to review some of the material.
If you find this approach useful, I'll try to get another one posted for next week.
I think it is
It is useful
yeah
OK...I'll post another one.
You should also buy now have a copy of an AP biology study guide. As we finish up human biology and start on Plant biology, you may want to take one of the example exams and score it, looking for two areas to improve: your test taking skills ( answering questions quickly, covering all of the material required), and subject matter areas in biology that you may need to review.
We're done for this morning.
I just took my diagnostic test
THank you for class!
Thanks for class!
thanks for class
We *will* go over the environment sections ahead of the actual AP exam and the rest of the class, so you'll get that material. Concentrate on any past/current material areas (microbio, inheritance, evolution, human anatomy) where you have problems.
See you in chat on Monday.
Bye! Have a great weekend!
bye every one